A Single Consortium View of Syntrophic Methane Oxidation by Environmental Archaea and Bacteria
Authors:
Victoria Orphan1* ([email protected], PI), Dan Utter1, Yongzhao Guo1, John Magyar1, Ranjani Murali4, Roland Hatzenpichler2, Gray Chadwick3, Hang Yu10, Shawn McGlynn6, Rex Malmstrom5, Danielle Goudeau5, Mark Ellisman7, Guy Perkins7, Tom Deerinck7, Jocelyn Richardson8, Sam Webb9
Institutions:
1California Institute of Technology; 2Montana State University; 3University of California–Berkeley; 4University of Nevada–Las Vegas; 5DOE Joint Genome Institute; 6Tokyo Institute of Technology; 7University of California–San Diego; 8SLAC National Accelerator Laboratory; 9Stanford Synchrotron Radiation Lightsource; 10Peking University
Abstract
The syntrophic anaerobic oxidation of methane (AOM) between methanotrophic archaea and sulfate-reducing bacteria is a critical microbially controlled process in the global methane cycle. These metabolic partnerships are diverse, representing several genera and species-level clades that co-occur in the same environment, segregated into individual two-member consortia believed to consist of a single archaeal and bacterial lineage. The high strain level diversity in sediments harboring AOM consortia frequently complicates the bioinformatic assembly of high-quality genomes from metagenomic datasets and the field currently lacks pure culture representatives for detailed physiological study.
Working with the Joint Genome Institute (JGI), Environmental Molecular Sciences Laboratory (EMSL), and the Stanford Synchrotron Radiation Lightsource (SSRL), this research team has been developing culture-independent strategies to investigate AOM consortia. Through support from the Facilities Integrating Collaboration for User Science program with JGI, the team developed flow cytometry sorting and sequencing protocols for individual AOM consortia from sediment in combination with click-chemistry enabled fluorescent tagging of translationally active microbes using a technique called BONCAT (BioOrthogonal Noncanonical Amino Acid Tagging). Application of the BONCAT-FACS method to these environmental methane-fueled microbial communities has enabled novel genomic insights into specific syntrophic partner pairings and hidden variation in metabolic potential (e.g., nitrogen fixation) among co-existing strains of methanotrophic archaea.
To complement these genome-guided analyses, researchers are working with SSRL and collaborators at National Center for Microscopy and Imaging Research to develop multimodal imaging for environmental AOM consortia that enables various combinations of fluorescence, electron, X-ray (µ-XRF, XANES), and secondary ion (NanoSIMS) imaging. As part of this effort, researchers have been testing SSRL’s upgraded 14-3 beamline for high resolution analysis of sulfur distribution and speciation in methane and sulfate-respiring consortia, enabling testing of hypotheses regarding the underlying syntrophic mechanism(s) of AOM. Collectively, these molecular, microscopy, and chemical/ isotopic imaging approaches are providing key insights into the physiology, ecology, and evolution of this globally important syntrophic partnership and importantly, offer a methodological roadmap that can be used with diverse microbial ecosystems.
Image
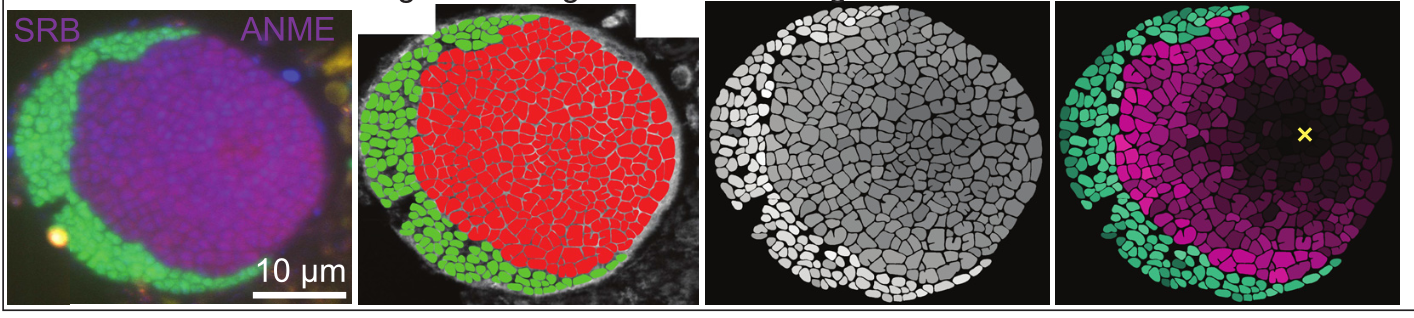
Overview of AOM Consortium Structure, NanoSIMS Data Acquisition and Single Cell Analysis of 15NH4 Assimilation Activity. Fractional abundance of 15N is calculated as 15N12C−/(15N12C−+14N12C−). Left to Right: Fluorescence in situ hybridization (FISH) image indicating phylogenetic identity of cells (green, bacterial probe; red, anaerobic methanotrophic archaea (ANME)-2 archaea-specific probe); 2nd Panel: Segmentation image showing sulfate-reducing bacteria (SRB) and ANME cells manually segmented based on observation of FISH and NanoSIMS data; 3rd Panel: Individual segmented cells shaded by their total 15N fractional abundance; 4th Panel: SRB and ANME cells scaled by minimum and maximum 15N enrichment values within the consortium. The yellow X marks the approximate minimum ANME cell activity several cell lengths away from their nearest syntrophic SRB partner. [Modified from He, X., et al., 2021. "Controls on Interspecies Electron Transport and Size Limitation of Anaerobically Methane-Oxidizing Microbial Consortia," MBio 12(3), 10-1128. Republished under Creative Commons license (CC BY 4.0)]
