Science Focus Area: Pacific Northwest National Laboratory
- Principal Investigator: Robert Egbert1
- Laboratory Research Manager: Vanessa Bailey1
- Co-Investigators:Joshua Elmore1, Pubudu Handakumbura1, Jason McDermott1, Bill Nelson1, Young-Mo Kim1, Ryan McClure1, Jennifer Brophy2, Adam Deutschbauer3, Adam Guss4, Carrie Harwood5, Devin Coleman-Derr6, Travis Wheeler7, Enoch Yeung8
- Participating Institutions:1Pacific Northwest National Laboratory, 2Stanford University, 3Lawrence Berkeley National Laboratory, 4Oak Ridge National Laboratory, 5University of Washington, 6University of California– Berkeley, 7University of Arizona, 8University of California Santa–Barbara
- KBase Collaboration: Improved protein annotation using machine learning, multi-omics data integration, and structural prediction
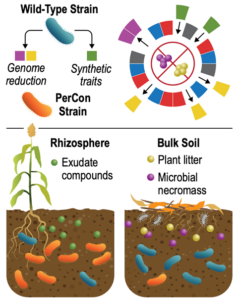
Persistence Control in the Rhizosphere. Conceptually, persistence control engineering involves manipulating catabolic pathways to shift the environmental niche of a microbe. This SFA aims to increase the environmental persistence in the rhizosphere of sorghum (left) and to decrease persistence under scavenging environments, such as plant detritus and microbial necromass (right). Led by the Pacific Northwest National Laboratory (PNNL), the Persistence Control SFA team is investigating how genome edits to non-model bacteria isolated from sorghum specifically alters fitness under laboratory conditions, in the presence of other microbes from the sorghum rhizosphere, and in the rhizosphere using laboratory and field measurements. The establishment of persistence control engineering principles will advance the responsible deployment of engineered microbial systems in the environment. [Courtesy PNNL]
Summary
The responsible deployment of engineered microbial rhizosphere communities offers tremendous opportunity to sustainably generate productive and stress-tolerant biomass cropping systems. A critical obstacle to realizing this vision is understanding the fundamental principles that drive plant growth promotion and environmental persistence for engineered functions in the rhizosphere. Specifically, strategies to boost the environmental persistence and impact of engineered functions among complex rhizosphere microbial communities and restrict growth away from the plant have not been established. By establishing persistence control as an engineering discipline to manipulate environmental niches, the Science Focus Area (SFA) aims to develop programmable control of engineered microbes through genome engineering to promote colonization of a target plant’s rhizosphere but to diminish in bulk soil when the plant stops growing or microbes migrate away from the rhizosphere.
Persistence control engineering relies heavily on a fundamental understanding of genes that contribute to fitness in rhizosphere and bulk soil environments. For selected engineered non-model rhizosphere bacteria, the SFA integrates existing annotations of gene function and high-throughput functional genomics data to enforce persistence control by altering the bacteria’s environmental niche through the introduction and removal of critical catabolic pathways. Using the bioenergy crop sorghum as a model rhizosphere system, the Persistence Control SFA team is developing genome editing tools for non-model rhizosphere bacteria, computational tools to predict persistence phenotypes in a community environment, small synthetic microbial communities as a testbed for persistence control escape, and an experimental framework to conduct rhizosphere science at scales from the laboratory bench to the field.
These efforts to understand and control rhizosphere persistence will lead to more thorough annotation of genes that contribute to microbial metabolism in the rhizosphere.
