Science Focus Area: Pacific Northwest National Laboratory
- Principal Investigator: Kirsten S. Hofmockel1
- Laboratory Research Manager: Jon Magnason1
- Co-Investigators: Nikos Kyrpides2, Hyun-Seob Song3, Steven Norberg4
- Participating Institutions: 1Pacific Northwest National Laboratory, 2U.S. Department of Energy Joint Genome Institute, 3University of Nebraska—Lincoln, 4Washington State University
- Project Website: pnnl.gov/projects/soil-microbiome
- Overview Brochure: Download PDF
- KBase Collaboration: Omics-enabled global gapfilling (OMEGGA) for phenotype-consistent metabolic network reconstruction of microorganisms and communities
Summary
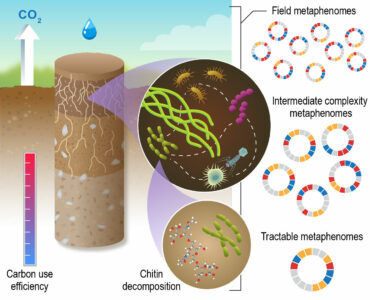
Soil Microbiome Metaphenome. The Soil Microbiome Science Focus Area, led by Pacific Northwest National Laboratory (PNNL), identifies molecular-level metabolic interactions governing decomposition, moisture effects on microbial phenotypes during decomposition, and their implications for carbon use efficiency. Work spreads across scales of biological and ecological complexity and uses complementary and synergistic network modeling approaches. Researchers are identifying how the genetic potential of individual microorganisms is expressed through interactions with other organisms and the environment, collectively composing a metaphenome. [Courtesy PNNL]
Pacific Northwest National Laboratory’s (PNNL) Soil Microbiome Science Focus Area (SFA) is using a cross-scale empirical and modeling approach to understand how enzymes, metabolites, and microbial consortia interact to control carbon cycling and sequestration. This approach will enable prediction of how these reaction networks and related functions shift in response to changing moisture regimes.
The SFA reconciles vast differences in scale and complexity by relating coarse metrics to molecular techniques and bridging organisms to metaphenomes to gain a predictive understanding of soil microbial communities and their function. Researchers use a genome-resolved, soil-derived, microbial consortium, paired with intermediate-scale modeling and empirical experiments, to identify biochemical and metabolic interactions that mediate soil microbiome functions identified under field conditions. Discoveries and outcomes from controlled experiments are being tested and validated in an irrigation field trial planted with tall wheatgrass, a potential bioenergy feedstock.
This SFA provides a comprehensive understanding of metabolic interactions and phenotypes from cellular to field scales. This understanding enables a mechanistic representation of microbial processes in multiscale models of integrated microbiome function. Results are providing much-needed information for discovering principles that translate from the lab to the field.
By providing a genomic understanding of the microbial metaphenomes measured throughout the rhizosphere and horizons of soil ecosystems, this work will help define biological paradigms that enable researchers to harness the soil microbiome’s functional capacity.
