National Laboratory Project: Sandia National Laboratories
- Principal Investigator: Joe Schoeniger1
- Co-Investigators: Kelly William1, Catharine Mageeney1, Jesse Cahill1, Yooli Light1, Michael Jewett2, Farren Isaacs3, Brady Cress4, Jennifer Doudna4
- Participating Institutions: 1Sandia National Laboratories, 2Northwestern University, 3Yale University, 4University of California–Berkeley
Summary
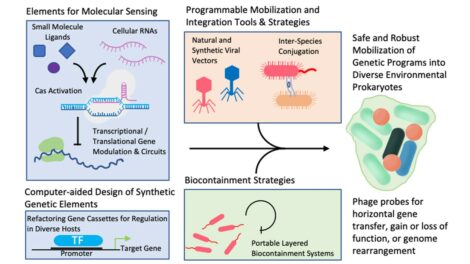
Microbial Community Composition. InCoGenTEC researchers are developing technologies to enable the study of changes in microbial community composition and genetics and the engineering of strong, multilayered biocontainment. Led by Sandia National Laboratories (SNL), the project’s technical themes include molecular tools for sensing and regulatory control, flexible genetic cassette design, bacteriophage vectors, and strategies for mapping and characterizing mobile genetic elements and controlling genetic mobility. [Courtesy SNL]
InCoGenTEC also aims to advance safe microbial gene editing by preventing the escape of heterologous/synthetic genetic content in modified microbes. To accomplish this unnecessary heterologous gene content is minimized and site-selective chromosomal integration of SGEs is used to avoid entraining them in natural genetic mobility processes revealed by mobility mapping analysis. Because DNA integrases can enable extremely efficient chromosomal integration, it could be possible to develop selection-free transformation methods that use native integration sites and can be applied in situ without the requirement for troublesome selection markers such as antibiotic resistance genes. In addition, biocontainment of SGEs and modified organisms is enabled by making activation of the SGE dependent on the host cell genetic content, and by implementing SGEs with modular biocontainment features, so that neither escaped genes or organisms can function. The SFA thus combines numerous mechanisms for molecular-context sensing and the regulation of gene expression and DNA recombination to enable stable strain-specific transformation of bacterial cells with enhanced biocontainment. Furthermore, engineered phage vectors that deliver SGEs that sense genetic material may be useful in experiments to detect escaped genes.
The team is designing their approach to enable the combination and modular use of these technologies with multiple layers of biocontainment security in a diverse range of non-model bacterial species found in environmental microbial communities. The SFA outcomes will advance research in understanding and editing microbiomes for biomanufacturing, biomass deconstruction, and soil and water microbiome ecology.
