Enhancing Biopreparedness Through a Model System to Understand the Molecular Mechanisms that Lead to Pathogenesis and Disease Transmission
Authors:
Margaret S. Cheung1,2* ([email protected], PI), David Pollock2,3, James Evans1,2, Weijun Qian1, Marat Valiev1, Olga Kuchar4, Arvind Ramanathan5, Melissa Peacock6, Omonike Olaleye7, Greg Morrison8, Zaida A. Luthey-Schulten9, Shannon Matzinger10
Institutions:
1Pacific Northwest National Laboratory; 2Environmental Molecular Sciences Laboratory; 3University of Colorado–Anschutz; 4Oak Ridge National Laboratory; 5Argonne National Laboratory; 6Northwest Indian College; 7Texas Southern University; 8University of Houston; 9University of Illinois Urbana-Champaign; 10Colorado Department of Public Health and Environment
Abstract
The science of biopreparedness to counter biological threats hinges on understanding the fundamental principles and molecular mechanisms that lead to pathogenesis and disease transmission. One vision to address this challenge is to create a powerful and user-friendly platform to elucidate the fundamental principles of how molecular interactions drive pathogen–host relationships and host shifts. Researchers will enable groundbreaking discoveries by integrating a wide range of structural, genomics, proteomics, and other advanced omics measurements, along with evolutionary and artificial intelligence predictions.
To make sure the system is applicable to real-world problems, it will be developed in the context of a tractable model system, the small, abundant, and accessible photosynthetic cyanobacteria and their constantly co-adapting viral pathogens, cyanophages. This model will maintain the system’s applicability to real-world problems and techniques, but the overall focus will be on elucidating general principles of detecting, assessing, and surveilling molecular interaction, adaptation, and coevolution that are system agnostic and therefore extensible to any other viral-host interaction.
The objectives are to (1) identify the molecular complexes that comprise the cyanobacteria redox macromolecular subsystem and how they dynamically change with bacteriophage infection in situ, using cryo-electron tomography; (2) profile regulatory changes during infection using proteomics, multiomics, and experimental validation, and integrate the data with in situ structures; (3) use genomics and metagenomics to determine environmental and population factors across time scales that impact the interactions between marine cyanobacteria and their cyanophage parasites, predicting the evolutionary origins of in situ structural and functional interactions, convergence and coevolution; and (4) develop a data integration and transformation platform that facilitates the integration of in situ, proteomic, and evolutionary measurements of molecular interactions to surveil diverse hosts and parasites in various environmental contexts.
This powerful and user-friendly platform will enhance connections between the often-siloed fields of structure, molecular phenotype, and evolutionary genomics that are key to biopreparedness, but in need of integration. The team will do this by building a navigation tool to facilitate the effective use of globally distributed experimental data for integrated analysis and predictive modeling. The impact of the project will be to develop, implement, and test a platform to assess host-pathogen molecular interactions, adaptation to hosts and host shifts, and coevolution between hosts and pathogens. A successful project outcome will transform researchers’ ability to study any host-pathogen interaction, encourage diverse community contributions, and gain fundamental insights into how proteins adapt to new contexts. This ability will be critical for designing early interventions to address future threats. Researchers will build surveillance training capability, aiming for a fair and equitable response to future pandemics and biothreats.
Image
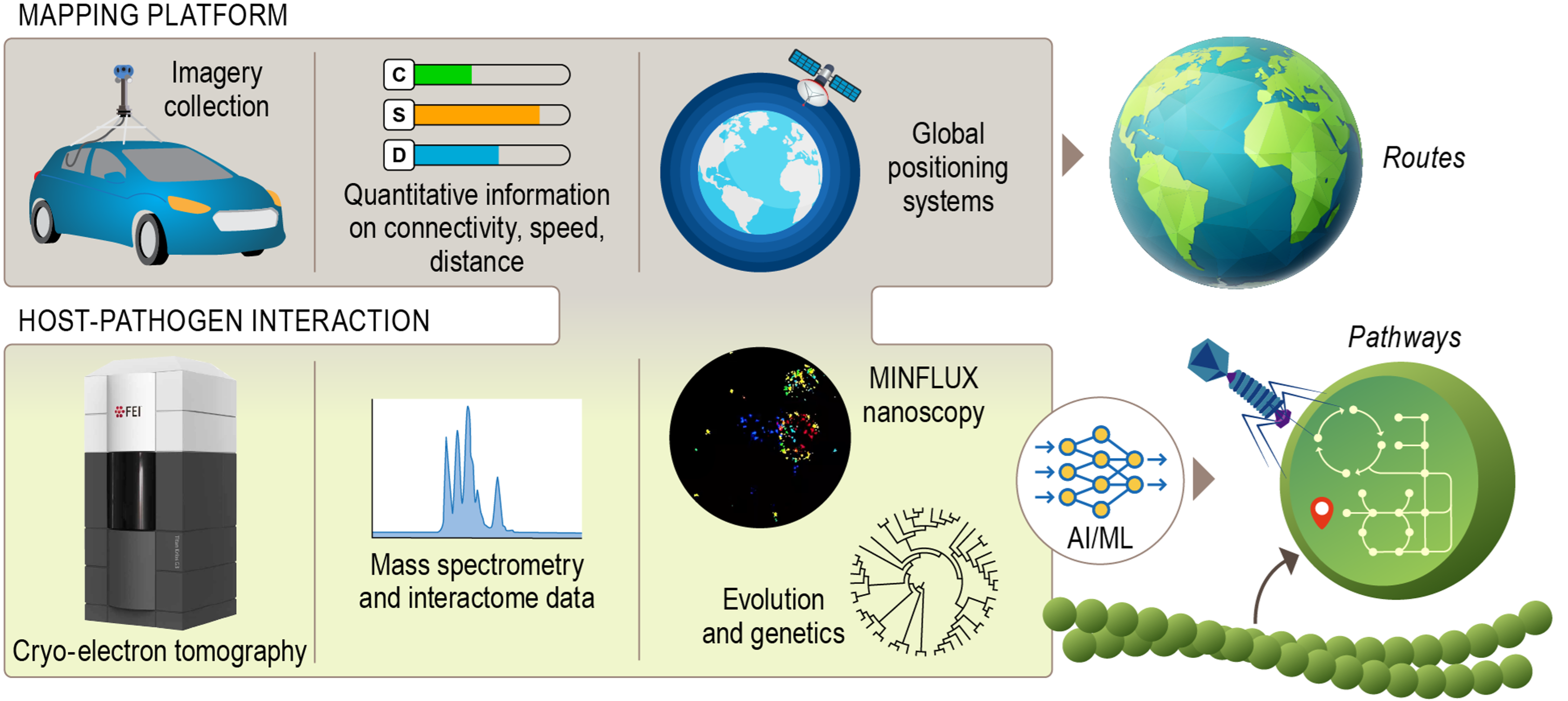
Platform for Mapping Host-Pathogen Interactions A platform for integrating multimodal datasets and modeling approaches toward targeting the host-pathogen interactions with equitable access to resources. [Courtesy Pacific Northwest National Laboratory]
