Getting Credit for Contributions in a Big Data World
Authors:
Elisha Wood-Charlson1* ([email protected]), Benjamin Allen3, Jason Baumohl1, Kathleen Beilsmith2, Joseph Bezouska2, David Dakota Blair4, John-Marc Chandonia1, Dylan Chivian1, Zachary Crockett3, Ellen G. Dow1, Meghan Drake3, Janaka N. Edirisinghe2, José P. Faria2, Jason Fillman1, Andrew Freiburger2, Tianhao Gu2, Prachi Gupta1, A. J. Ireland1, Marcin P. Joachimiak1, Sean Jungbluth1, Roy Kamimura1, Keith Keller1, Dileep Kishore3, Dan Klos2, Filipe Lui2, David Lyon1, Cody O’Donnell1, Mikaela McDevitt1, Christopher Neely1, Erik Pearson1, Gavin Price1, Priya Ranjan3, William Riehl1, Boris Sadkhin2, Samuel Seaver2, Alan Seleman2, Gwyneth Terry1, Pamela Weisenhorn2, Ziming Yang4, Shinjae Yoo4, Sijie Xiang1, Qizhi Zhang2, Shane Canon1, Paramvir S. Dehal1, Robert Cottingham3, Christopher S. Henry2, Adam P. Arkin1 (PI)
Institutions:
1Lawrence Berkeley National Laboratory; 2Argonne National Laboratory; 3Oak Ridge National Laboratory; 4Brookhaven National Laboratory
URLs:
Goals
The DOE Biology Knowledgebase (KBase) is a knowledge creation and discovery environment designed for both biologists and bioinformaticians. KBase integrates a large variety of data and analysis tools, from DOE and other public services, into an easy-to-use platform that leverages scalable computing infrastructure to perform sophisticated systems biology analyses. KBase is a publicly available and developer extensible platform that enables scientists to analyze their own data within the context of public data and share their findings across the system.
Abstract
Many of the concerns researchers have around sharing data include knowledge barriers, reuse concerns, and disincentives (Gomes et al. 2022). KBase already addresses components of knowledge barriers through its outreach strategy (e.g., in-person and virtual training sessions, documentation, and robust hands-on activities) and reuse concerns through the basic functionality of the platform (e.g., provenance, interoperability, and reproducibility) (Arkin et al. 2018). KBase is now working to address disincentives: concerns around getting scooped, the time it takes to curate and share data, and lack of clarity around the rewards that come from embracing open science and good data management.
Behind much of this is the fundamental tenet that neither the current culture of science nor the publishing infrastructure value data outside of a publication. In partnership with several efforts across BER and the publishing world, KBase is establishing its platform as a change agent focused on “getting and giving credit to data” (Wood-Charlson et al. 2022). Leveraging Persistent Identifiers (PIDs), group members are developing linkages to/from and within KBase that support data management best practices and ensure credit is retained. An example user (PID: ORCID) research workflow:
• Collect samples and assign them International General Sample Numbers (PID: IGSN),
• Submit sample metadata (Standard: MIxS) to the National Microbiome Data Collaborative (NMDC) Sample Submission Portal,
• Send sample material to the DOE Joint Genome Institute ( JGI) for sequencing,
• Send sample material to the Environmental Molecular Sciences Laboratory (EMSL) for Fourier-transform ion cyclotron resonance (FTICR) mass spectrometry analysis, and
• Submit geochemistry measurements on those samples to Environmental System Science Data Infrastructure for a Virtual Ecosystem (ESS-DIVE).
Leveraging the (in development) Data Transfer Service (DTS), the user could request sample metadata from NMDC, sequence data from JGI, and FTICR data from EMSL to do a global community analysis and build community models. When the user is ready to publish their data and reproducible analyses, the KBase “Credit Engine” provides the user’s workflow (“Narrative”) with a dataset DOI that captures (Ireland and Wood-Charlson 2023):
1. Important credit metadata [e.g., authors/contributors (PID: ORCID), funder information (PID: Research Organization Registry, ROR)]
2. Citations for the funded proposal/data management plan (PID: DOI), samples (PID: IGSN), public data (PID: DOI, when available), and tools (DOI, typically a publication) that contributed to the analysis.
The KBase DOI is registered by Office of Scientific and Technical Information and submitted to DataCite, which links the now FAIR (findable, accessible, interoperable, and reusable) Narrative to the broader publishing infrastructure, and shared back to the ESS-DIVE project. These connections enable KBase to start tracking and reporting the reuse of shared data.
Image
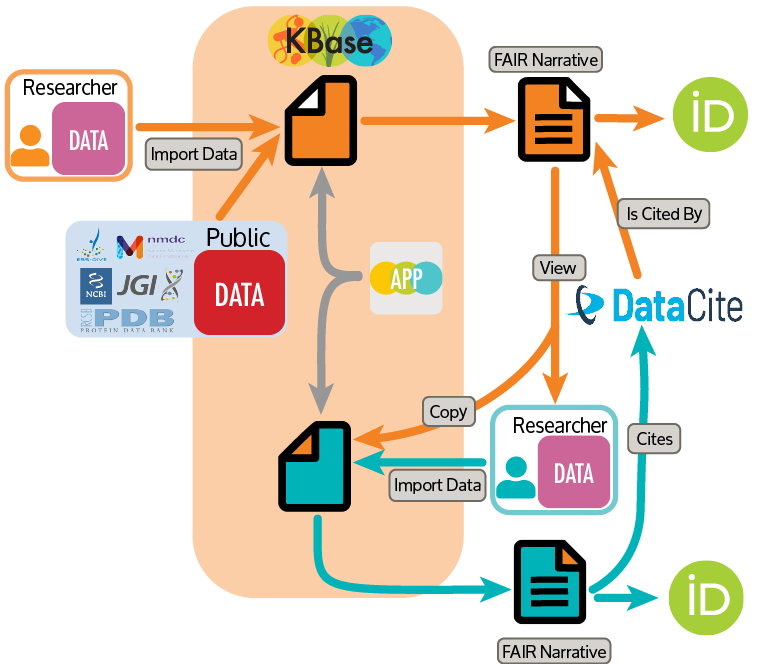
DOE Biology Knowledgebase (KBase). Researchers using KBase can automatically get credit for their research by requesting a DOI for their KBase Narrative associated with a publication. [Courtesy KBase Credit Engine Team]
References
Arkin, A. P., et al. 2018. “KBase: The United States Department of Energy Systems Biology Knowledgebase,” Nature Biotechnology 36(7), 566–9. DOI:10.1038/nbt.4163.
Gomes, D. G. E., et al. 2022. “Why Don’t We Share Data and Code? Perceived Barriers and Benefits to Public Archiving Practices,” Proceedings of the Royal Society B: Biological Sciences 289, 20221113. DOI:10.1098/rspb.2022.1113.
Ireland, A. J., and E. M. Wood-Charlson. 2023. “KBase Credit Metadata Schema.” Accessed April 2024. KBase. DOI:10.25982/1984203.
Wood-Charlson, E. M., et al. 2022. “Ten Simple Rules for Getting and Giving Credit for Data,” PLoS Computational Biology 18(9), e1010476. DOI:10.1371/journal. pcbi.1010476.
Funding Information
This work is supported as part of the Genomic Science program of BER. The DOE Systems Biology Knowledgebase (KBase) is funded by the DOE Office of Science, BER program under DE-AC02-05CH11231, DE-AC02-06CH11357, DE-AC05-00OR22725, and DE-AC02-98CH10886.
