Novel In Vivo Visualization of Bioenergy Metabolic and Cellular Phenotypes in Living Woody Tissues
Authors:
Andrew Groover2* ([email protected]), Al Ingold1, Ruipeng Guo1, Rajesh Mennon1, Reed S. Sorenson1, Gayatri Mishra1,2, Dasmeet Kaur3, Chang-Jun Liu3, Leslie E. Sieburth1 (PI)
Institutions:
1University of Utah; 2U.S. Forest Service, Northern Research Station; 3Brookhaven National Laboratory
Abstract
This research team is developing new approaches for live cell imaging within the wood-forming tissues of trees enabling deeper understanding of developmental and physiological processes underlying wood formation and function directly in woody bioenergy feedstocks. A primary challenge is that the dividing and differentiating cells of interest are embedded under thick layers of light-scattering bark tissues and are well beyond the working distances of traditional light microscopes.
The group is developing two types of microendoscopic, implantable imaging probes to access and visualize wood-forming tissues: miniscopes and fiber optic cannulas. These microscopy approaches are being tested by applying them to the following problems relevant to bioenergy feedstock development in poplar: (1) analysis of lignification and impacts of the altered cell wall polysaccharides on lignin formation; (2) vessel element differentiation and the impact of abscisic acid levels on vessel cell properties; and (3) fiber development in both tension-wood-inducing and normal growth conditions.
Previously, the group reported progress in fabricating and using embedded optical probes to carry out live cell imaging in poplar stems. This poster reports improvements to epifluorescence miniscopes fitted with implantable Gradient Index (GRIN) lenses inserted to reach internal tissues with minimal tissue damage. This approach has the advantage of directly rendering images and video in real time. It can be used in combination with fluorescent probes and dyes compatible with a range of different excitation and emission filters. However, miniscope resolution is challenged by light scattering in woody stems with complex autofluorescence.
The group is using ring deconvolution microscopy (Kohli et al. 2023), a technique for computational aberration with a calibration image of randomly distributed fluorescent microspheres. The second approach uses computational cannula microscopy with an individual cannula or an optrode array (Guo et al. 2023), which can be modified to enable hyperspectral imaging. This approach has the advantages of smaller diameter probes (0.22 mm) and larger fields of view (0.20 mm). However, in contrast to the miniscope, machine learning–based image processing algorithms are necessary to convert spatially scrambled fluorescent signals into images. The group implemented a generative adversarial convolutional neural network that surpasses the capabilities of the prior U-Net architecture (Isola et al. 2018). This advancement enables reconstructed images of living poplar branches, which display low contrast and dense structures and capture of dynamics at a cellular level. Finally, researchers will discuss opportunities to extend live cell imaging capabilities for woody plant bioenergy applications.
Image
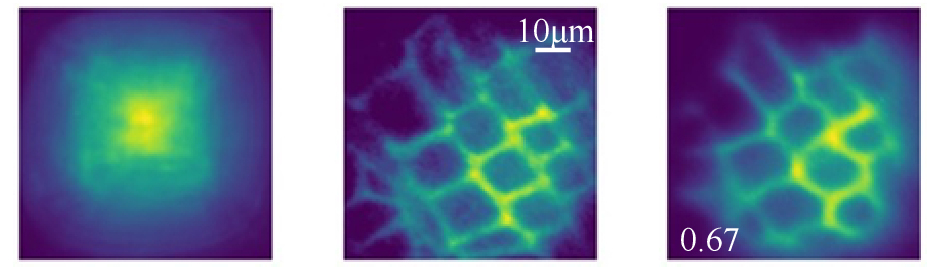
Imaging Sections of Stems. Left: A cannula directs excitation light into the plant and collects emitted fluorescence, which is then recorded by a camera. Middle: Ground-truth image obtained using a conventional reference microscope. Right: Reconstructed image after computational post- processing of the recorded image. [Adapted with permission from Guo, R., et al. 2023. "Overcoming the Field-of-View to Diameter Trade-Off in Microendoscopy via Computational Optrode-Array Microscopy," Optica Express 31(5). DOI:10.1364/OE.478314. © 2024 Optica Publishing Group under the terms of the Optica Open Access Publishing Agreement]
References
Guo, R., et al. 2023. “Overcoming the Field-of-View to Diameter Trade-Off in Microendoscopy via Computational Optrode-Array Microscopy,” Optics Express 31(5), 7505–14. DOI:10.1364/OE.478314.
Isola, P., et al. 2018. “Image-to-Image Translation with Conditional Adversarial Networks,” arXiv. DOI:10.48550/ arXiv.1611.07004.
Kohli, A., et al. 2023. “Ring Deconvolution Microscopy: An Exact Solution for Spatially-Varying Aberration Correction,” arXiv. DOI:10.48550.arXiv.2206.08928.
Funding Information
This research was supported by the DOE Office of Science BER program, under award number DE-SC0021996, and Interagency Agreement Number 89243021SSC000074 to AG.
