Understanding Plant–Environmental Interactions Using Single-Cell Approaches
Authors:
Benjamin Cole2* ([email protected], PI), Karen Serrano1, Margot Bezrutczyk2, Danielle Goudeau2, Thai Dao1,2, Rex Malmstrom2, Henrik Scheller1
Institutions:
1Joint BioEnergy Institute; 2DOE Joint Genome Institute
Abstract
Biomass derived from plant feedstocks is a renewable and sustainable energy resource, but these resources are vulnerable to environmental stress such as water and nutrient limitations. Understanding how cells work independently and in concert to regulate plant responses to their environment, including their surrounding microbial community, as well as abiotic stress will be crucial to improving their performance.
This group has applied several cutting-edge, single-cell, and spatially resolved transcriptome sequencing approaches to several plant species and is constructing a comprehensive single-cell resource for plants to better understand the complexity behind environmental responses among diverse cell types. In particular, the group has leveraged both single-nuclei and spatial transcriptomics to profile interactions between plants and arbuscular mycorrhizal fungi (AMF) using the Medicago truncatula and Rhizophagus irregularis system. The team developed a cross-kingdom transcriptome map for this crucial symbiosis, profiling both plant and fungal expression patterns. Team members are also working toward establishing more precise spatial omics tools to profile tissues from bioenergy grasses (including sorghum, switchgrass, and Brachypodium) reacting to AMF colonization, as well as physical stresses including nutrient deprivation and osmotic stress. The team hopes to build a multi-species model of cell type–specific environmental responses.
Image
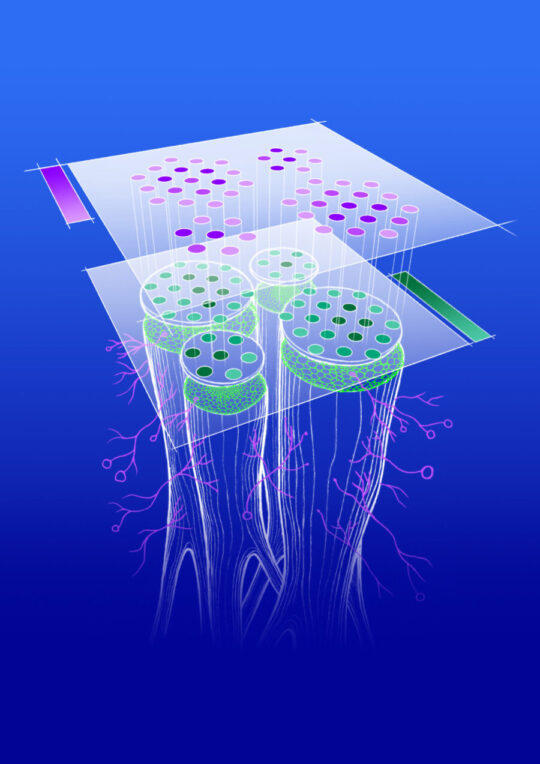
Spatial Co-Transcriptomics Reveals Discrete Stages of the Arbuscular Mycorrhizal Symbiosis. This 2024 publication shows stylized Medicago root colonized by arbuscular mycorrhizal fungi and profiled using spatial transcriptomics technology. [Republished under Creative Commons License 4.0 from 2024. "Symbiosis in Time and Space," Nature (CC BY 4.0)]
Funding Information
This work was supported by the U. S. Department of Energy, Office of Science, BER program, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and DOE. This work was also supported by an Early Career Research program to B.C., the Laboratory Directed Research and Development program at Lawrence Berkeley National Laboratory, and by the DOE Joint BioEnergy Institute.
