Utilizing Metaomic and Chemical Imaging Approaches to Explore the Dynamics of Nutrient Exchange and Environmental Adaptation Mediated by Plant–Fungal Symbiosis
Authors:
Hui-Ling Liao1* ([email protected]), Haihua Wang1, Kaile Zhang1, Ryan Tappero3, Tiffany Victor3, Rytas Vilgalys4, Kerrie Barry5, Keykhosrow Keymanesh5, Sravanthi Tejomurthula5, Igor V. Grigoriev5, William R. Kew6, Elizabeth K. Eder6, Carrie D. Nicora6, John Cliff 6, Jeremy Bougoure6, Dehong Hu6, Sarah Leichty6, Edward Brzostek7, Jennifer Bhatnagar2 (PI)
Institutions:
1University of Florida; 2Boston University; 3Brookhaven National Laboratory;4Duke University; 5DOE Joint Genome Institute; 6Pacific Northwest National Laboratory; 7West Virginia University
URLs:
Abstract
Interactions among terrestrial free-living microorganisms, root symbiotic microbes, and plants play a crucial role in modulating biogeochemical cycling within ecosystems. However, the dynamic mechanisms of these interactions and their responses to changing environmental conditions, such as soil nutrient status and stresses are poorly understood. Consequently, terrestrial nutrient fluxes in ecosystem models often overlook plant–symbiont–microbe interactions and cannot accurately reflect the impacts of environmental changes on their functions. Using pine trees as a model, researchers employ advanced metaomic, NanoSIMS and X-ray fluorescence (XRF) tools to uncover the mechanisms by which plants, ectomycorrhizal fungi, and free-living soil microorganisms interact to shape nutrient exchange and decomposition processes.
This experiment was established in a phytotron-based synthetic ecosystem for pine and its soil microbiomes. Treatments include the combination of soil sources harboring diverse soil biomes, elevated carbon dioxide (CO2) levels, soil organic matter (SOM), inorganic nitrogen (N), iron (Fe), and pine-associated ectomycorrhizal fungi (EMF).
Findings from the subset of these treatment combinations indicated that an enriched abundance of EMF facilitated soil molecular and extracellular enzyme activities, involved in soil carbon (C), N, and Fe processes. NanoSIMS images revealed the ability of EMF-extended hyphae to acquire 15N from 15N- labeled SOM. Metaomic studies demonstrated that elevated CO2 levels enhanced EMF pathways involved in symbiosis- mediated SOM decomposition and N transfer from the soil to the host plants.
However, the treatments with inorganic N or Fe compromised symbiotic C/N processes; such negative impacts can be mitigated by enhancing the diversity of the EMF community. The ongoing study integrating data from metaomic analyses and XRF will discuss the interactive effects of SOM, N, and Fe on free-living microorganisms and EMF-mediated nutrient fluxes.
The team’s next goal is to integrate these data into an ecosystem model to represent plant–microbe interactions under global change scenarios and their feedback to ecosystem-level nutrient fluxes. The team anticipates that this refined model will better capture the impacts of plant–microbe interactions on biogeochemical cycling in changing environments and inform forest management practices.
Image
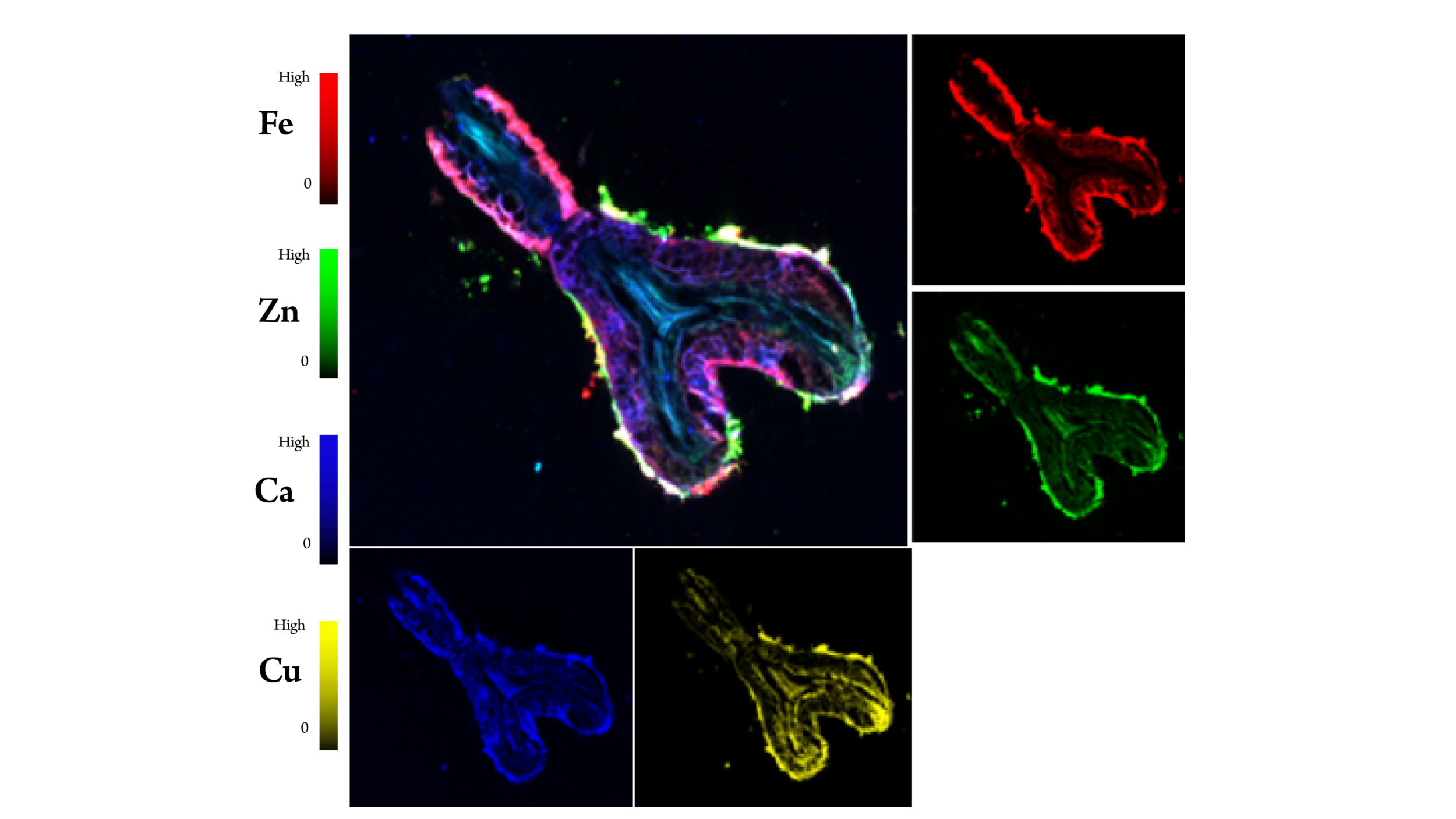
X-Ray Fluorescence. Images indicate the uptake of iron (Fe), zinc (Zn), calcium (Ca), and copper (Cu) in a mycorrhizal root tip of Pinus taeda inoculated by Suillus cothurnatus under conditions of relatively low inorganic nitrogen and high soil organic matter. [Courtesy University of Florida and Brookhaven National Laboratory]
