Predicting and Modeling Protein-Protein Complexes at Large-Scale with Deep Learning
Authors:
Mu Gao1* ([email protected]), Davi Nakajima An1, Jerry M. Parks2, and Jeffrey Skolnick1
Institutions:
1Georgia Institute of Technology; and 2Oak Ridge National Laboratory
Goals
With the advances in next-generation sequencing technologies, the number of sequenced genomes is growing exponentially. This has resulted in a bottleneck for the translation of sequence information into functional hypotheses about each gene. Current gene annotation technologies are primarily based on evolutionary inference by sequence comparison; however, many proteins in a proteome remain uncharacterized. To address this challenge, this collaborative team is developing a suite of novel high-performance-computing (HPC), deep-learning methods that predict protein structures and interactions at unprecedented accuracy, making use of the Summit supercomputer at the DOE leadership computing facility at the Oak Ridge National Laboratory. The combination of deep learning, HPC, and structural-based analysis will help to understand molecular mechanisms of protein functions, and enable rapid, accurate prediction of gene function on a genomic scale, such as novel protein-protein interactions important to life.
Abstract
One key observation of proteins in a living cell is that they usually interact with each other to carry out their biological functions. The identification and characterization of these protein-protein interactions are therefore critical to understanding life. Very recently, researchers proposed a deep learning–based approach for the identification of protein-protein interactions. The approach, AF2Complex, is built on the success of AlphaFold 2. AF2Complex extends the idea of structure modeling of a single protein sequence to a complex made of multiple sequences and further predicts protein-protein interactions by using the confidence of its structural modeling. While AF2Complex have been successfully benchmarked in multiple tests including 7,000 protein pairs from the bacteria E. coli, it is important to demonstrate its usefulness by applying it to address some real-world problems. For this purpose, team members investigated the pathway leading to the folding and assembly of outer membrane proteins (OMP) in E. coli as a proof-of-concept illustration of approach. OMPs serve an essential functional role such as nutrients exchange with their living environment. The making of these barrel-like OMP proteins is an elaborate process starting within the cytoplasm, where they are first manufactured by ribosomes. Coming out of the ribosomes are nascent, still largely unfolded peptide chains that must subsequently cross the inner membrane, travel through the periplasmic space, and finally land at their destination: the outer membrane. To ensure a successful journey, many other proteins provide vital help by forming functional protein complexes. However, they are challenging for experimental characterization because many of them are membrane proteins, and the interactions are often transient. By applying the AF2Complex workflow established at Summit to several key proteins in the OMP biogenesis pathway, researchers have identified their functional partners within the top 1% ranking of ~1,500 proteins screened for PPIs per query. Thanks to high confidence structures underlying the top predictions, one can understand many experimental phenomena, particularly in vivo site-directed photo cross- linking data. For example, cross-linked products found from the translocon SecYEG or the β-barrel assembly machine (BAM) supercomplexes may be explained by direct physical interactions revealed in predicted structures (Figure 1). An unexpected, biologically important interaction has been identified between the enzyme DsbA and chaperon PpiD, which is associated with the SecYEG translocon. Moreover, previously speculated conformations are captured for SurA and BepA. Most importantly, these revealing atomic structures of various supercomplexes suggest mechanistic hypotheses for various steps of the OMP biogenesis pathways.
Image
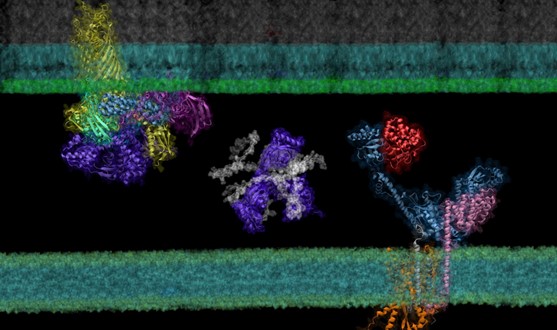
Figure 1. Super protein complexes in the outer membrane biogenesis pathway in E. coli identified and modeled by screening the envelopome of E. coli with AF2Complex.
References
Gao, M., et al. 2022. “AF2Complex Predicts Direct Physical Interactions in Multimeric Proteins,” Nature Communications 13, 1744. https://www.nature.com/articles/s41467-022-29394-2.
Gao, M., et al. 2022. “Deep Learning-Driven Insights into Super Protein Complexes for Outer Membrane Protein Biogenesis in Bacteria,” eLife 11, e82885. https://elifesciences.org/articles/82885.
Funding Information
This research was supported by the DOE Office of Science, Office of Biological and Environmental Research (BER), grant no. DE-SC0021303, and the Advanced Scientific Computing Research (ASCR) Leadership Computing Challenge (ALCC) program. The research used resources provided by the Leadership Computing Facility at Oak Ridge National Laboratory, National Energy Research Scientific Computing Center at Berkeley, and the Partnership for an Advanced Computing Environment (PACE) at Georgia Tech.
